I have been kicking around the idea of setting up an online catalog of lichen photobionts for years before I started doing this. The main impetus to finally start was this paper:
Fernández-Martínez, M., de los Ríos, A., Sancho, L., & Pérez-Ortega, S. (2013). Diversity of Endosymbiotic Nostoc in Gunnera magellanica (L) from Tierra del Fuego, Chile Microbial Ecology DOI: 10.1007/s00248-013-0223-2
I have been interested in the relationships between lichenized Nostoc and those that form symbioses with ferns, cycads, liverworts and the flowering plant Gunnera for a long time, but my efforts to address the issue were hampered by inadequate sampling and others who had better sampling used a genetic marker with a complex history that made such comparisons difficult. Finally, here was a paper with extensive sampling across the range of a plant host of Nostoc who used the same genetic marker that has been widely adopted in work on lichen photobionts. Unfortunately, while the paper has a lot of interesting things to say about the Nostoc-Gunnera symbiosis (genetically monomorphic within individuals, lots of variability among individuals, reduced symbiont diversity in recently deglaciated areas, etc), they included very few lichen photobionts in their analyses, so I wasn’t able to get the answers to the questions I’m interested in from the paper.
Now that I’ve developed a decent phylogentic framework for symbiotic Nostoc, it should now be possible to address these questions. Here is how the G. magellanica symbionts fit in:
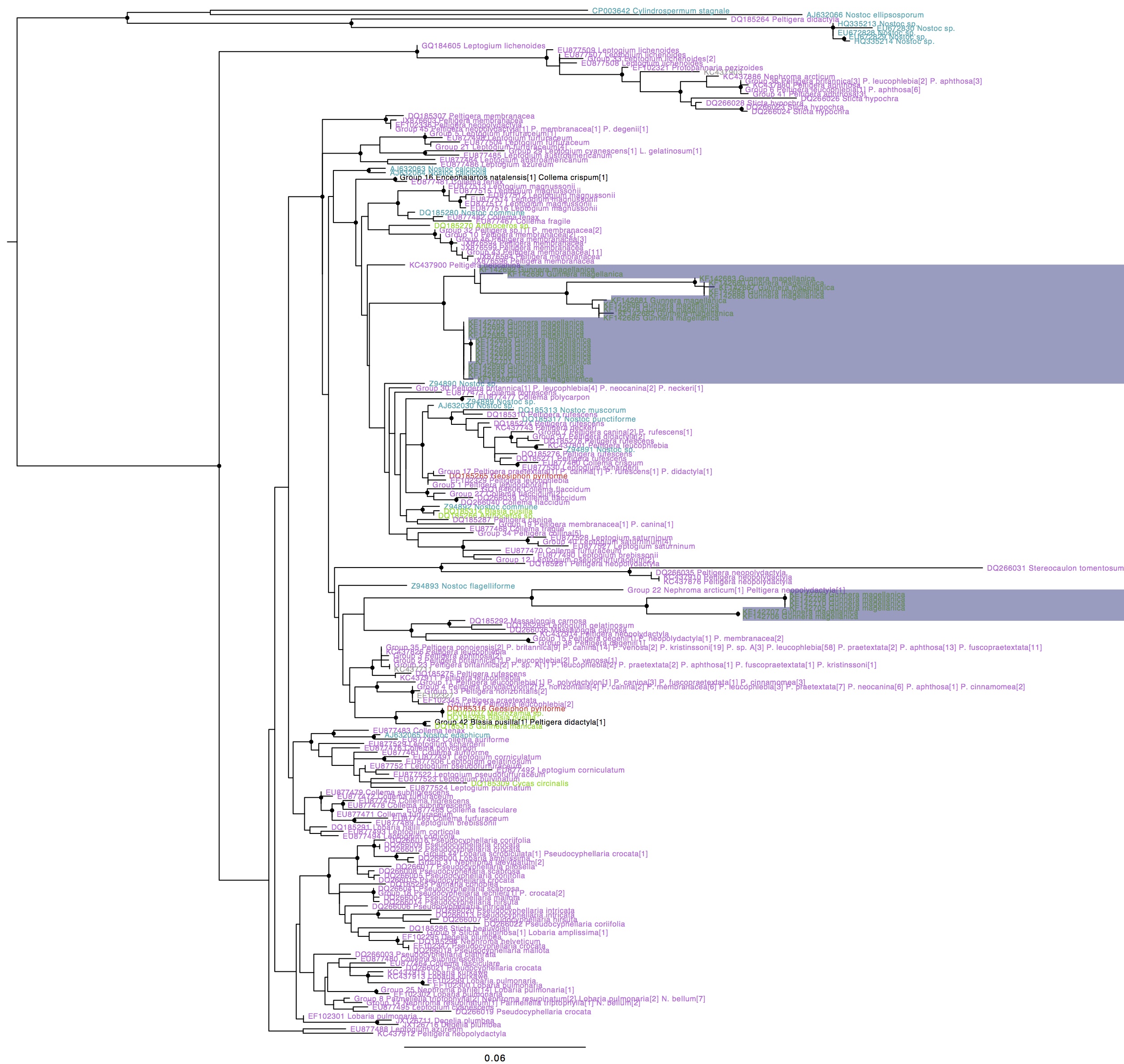
Nostoc rbcX phylogeny with Gunnera magellanica symbiont hilighted, coloured by type of association (purple: lichen photobionts, green: plant symbionts, blue: free-living, red: fungal endosymbiont). Names in black indicate genotypes found in more than one group. Circles on internal nodes indicate aLRT ≥0.9.
A few things about this tree are interesting. For one, G. magellanica symbionts form two well-supported lineages to exclusion of all other strains. This contradicts the results from the paper, where two G. magellanica symbiont haplotypes did not group with the others and where one lichen photobiont was nested within one of the G. magellanica symbiont clusters, though resolution and support were low for these nodes in their tree. Secondly, the G. magellanica symbionts do not group with any other plant symbionts, including the other Gunnera symbiont. Thirdly, the G. magellanica symbionts are on relatively long branches, suggesting that the evolutionary rate is higher in this lineage.
One major note of caution are in order when interpreting this tree, however: all of these G. magellanica symbionts were collected in the southern tip of South America, while the vast majority of the sampling of other lineages, including many of the plant symbionts, is from the northern hemisphere. Indeed, several of the symbionts of tropical plants were isolated from botanic gardens in Europe, well outside of the native range of the plants. It is certainly possible that lichens and other plant hosts from South America would associate with some of the same Nostoc strains isolated from G. magellanica.
So, in conclusion, it’s fair to say that these data support the notion of frequent host shifts between lichens and plants in the evolutionary history of Nostoc, but that there is no evidence that the same strains of Nostoc routinely form symbionses with lichens and with Gunnera. However, more geographically appropriate sampling may provide this evidence in the future.
Methods:
Sequences KF142679 to KF142710 were downloaded from genbank and added to the Nostoc rbcX dataset. These 32 haplotype sequences represent 110 specimens, but it is not clear how many specimens each haplotype represents. Details of the analysis can be found here. Data files are here.
Heath OBrien (2013). Gunnera symbionts do not cluster with lichen photobionts PhotobiontDiversity.wordpress.com : http://dx.doi.org/10.6084/m9.figshare.726140


Would sampling of Gunnera-Nostoc outside of its normal South American range be interesting/helpful, such as from botanical gardens?
I think previously you started to look at genome architecture of cyanobacterial strains. Are there enough genomes available now for you to compare Gunnera-Nostoc to other Nostocs? I’m not saying that’s where the answer is, just curious if there are noticeable differences in genome content between Nostocs that jump hosts, and those that don’t jump hosts.
The Next-gen genomics revolution seems to have completely passed the world of freshwater/terrestrial cyanobacteria by. There aren’t any Nostoc sequences in the NCBI WGS archive and last I checked, there was only one fully sequenced strain. Definitely an area with a lot of potential.
As for Gunnera from botanic gardens, there is one sequence from one in Germany in the tree. It groups with other plant symbionts and with one Peltigera photobiont. It would be interesting if they are forming novel symbioses in these introduced habitats. Of course there could be differences between Gunnera species as well.
Pingback: Another perspective on diversity of symbiotic cyanobacteria: 16S | PhotobiontDiversity