Having beaten the phylogeny of symbiotic cyanobacteria into submission in my previous post, I am now tackling the green algae. My plan was to start with a big-picture analysis of 18S ribosomal RNA sequences, but my initial blast search returned over 10,00o 454 reads from metagenomic projects which was a lot more “environmental isolate XXX” than I felt like dealing with. Besides, I don’t know that I could add much to this recent overview. Therefore, I am going to focus on the most important lineage of lichenized algae: Trebouxia. There have been a large number of studies that have obtained photobiont ITS sequences from a variety of Trebouxia associated lichens, so these are the data that I looked at.
The methods are the same as the ones that I described in detail previously for Nostoc ITS sequences. Briefly, I used two ITS sequences (T. impressa JN204819 and T. arboricola JQ993781) as queries to identify all homologous (E-value <= 1e-100) sequences in the nt database. Sequences were aligned with MAFFT, duplicate sequences were removed with MetaPIGA, alignment positions corresponding to gaps in the references sequence (T. arboricola JQ993758) were removed with trimal, and phylogenetic relationships were inferred with PhyML.
This procedure produced a tree with 794 taxa representing 1840 Trebouxia ITS sequences. The actual number of Trebouxia associated lichens that have been sequenced is much higher than this because many authors only deposit representative sequences of each haplotype that they obtained. At some point I will dig into the papers where this has been done to extract the real numbers, but I have not done so yet.
For now, I am going to focus on the taxonomy of the algae. I will leave a discussion of the host-association patterns for a future post. Here is the Trebouxia ITS phylogeny color-coded by species (tree file can be found here):
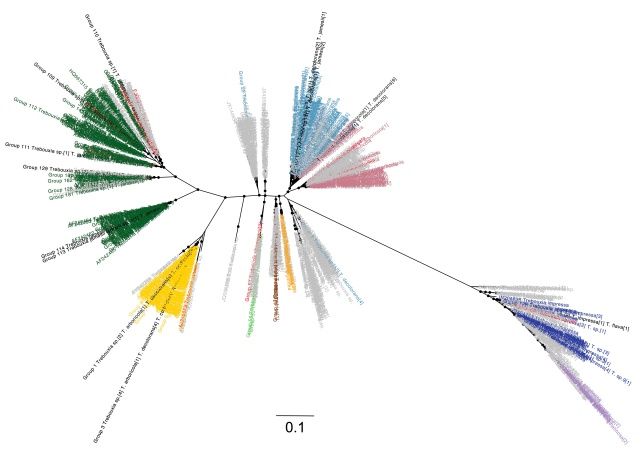
Trebouxia ITS phylogeny color-coded by species (dark green: T. jamesii, yellow: T. corticola, light green: T. incrustata, brown: T. asymmetrica, ornage: T. gigantea, purple: T. gelatinosa, dark blue: T. impressa, light red: T. arboricola, light blue: T. decolorans, dark red: other, grey: T. sp.). Sequences recovered from multiple named species are in black. Black circles indicate aLRT support > 0.9
With a few exceptions, sequences from named algae tend to cluster very well. T. gelatinosa (purple) is nested within T. impressa (dark blue), though given the long branch separating these two species from all of the others, I don’t entirely trust the rooting of this clade. T. jamesii (dark green) is a very heterogeneous group as has been recognised previously. A number of photobionts that group with T. decolorans (light blue) have been identified as T. arboricola (light red). Three major lineages have no named members (except for some presumably misidentified T. decolorans sequences).
In addition to the differentially coloured species, there are several additional species names that are represented by a small number of sequences, all of which are colored dark red in the tree. T. australis, T. brindabellae, T. showmanii and T. usneae are each found in distinct clusters and are likely to represent additional good species. T. australis and T. brindabellae are both in clusters near the base of one of the T. jamesii clades (dark green). Two T. showmanii sequences form the sister group to T. incrustata (light green). T. usneae forms a distint lineage with a misidentified T. corticola sequence sister to the T. corticola lineage (yellow). All other rare species are deeply nested within other common species and appear not to be distinct. These include T. potteri which is nested within T. impressa (dark blue), T. aggregata and T. crenulata which are nested within T. arboricola (light red) and T. simplex, which includes six sequences that are identical to T. jamesii (black) and two other sequences that are nested within one of the T. jamesii clades (dark green). T. flava is identical to a T. impressa sequence and is coloured black in the tree.
In conclusion, >1840 Trebouxia ITS sequences that have been obtained from lichens cluster into about 24 distinct species, 13 of which appear to have suitable named representatives in the database. Two of the T. jamesii clusters have been given the provisional names T. “vulpinea” and T. “letharii” but it looks like at least three additional names are needed for this group.
That’s it for now. In my next post I will map host information onto this tree.
Heath OBrien (2014). Green Algal Photobionts: Trebouxia. PhotobiontDiversity.org: http://dx.doi.org/10.6084/m9.figshare.703888


Pingback: A preliminary look at host association patterns in Trebouxia | PhotobiontDiversity